Researchers Develop ELI Trifocal Microscope for Precise Target cryo-lamellae Preparation
In a recent study, Prof. Dr. SUN Fei's group at the Institute of Biophysics (IBP) of the Chinese Academy of Sciences (CAS) developed an advanced precise and efficient cryo-FIB fabrication technique with the name ELI-TriScope and applied this technique to study the 3D in situ structure of the human centriole.
In ELI-TriScope, an electron (E) beam, a light (L) beam and an ion (I) beam are precisely adjusted to the same focal point; as a result, cryo-FIB milling can be accurately navigated by monitoring the real-time fluorescence signal of the target molecule (Fig. 1).
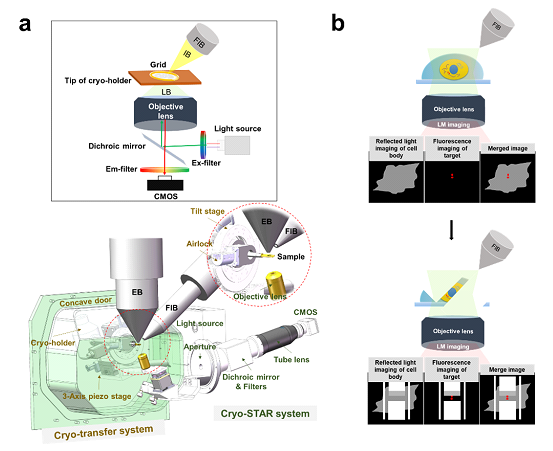
Figure 1. Design and principle of ELI-TriScope. (a) Schematic diagram and design drawing of ELI-TriScope in its operational mode. Each part of the system is labelled and described. IB, ion beam. LB, light beam. EB, electron beam. FIB, focused ion beam. (b) Working principle of ELI-TriScope. The ion beam and light beam are simultaneously focused on the area of interest of the cryo-specimen. The reflected light image recorded by the camera displays the cell body, and the target signal in the fluorescence image is used to navigate cryo-FIB milling.
Furthermore, they prepared 72 high-quality cryo-lamellae of HeLa cells with a high success target rate (~91%) by using ELI-TriScope, and collected 75 cryo-ET tilt series of cryo-lamellae and study the in situ structure of the human centriole.
More importantly, they observed multiple components of human centrioles in their native states. With a preliminary STA procedure, it could resolve the single protofilaments of MTTs and observe their longitudinal periodicity (Fig. 2).
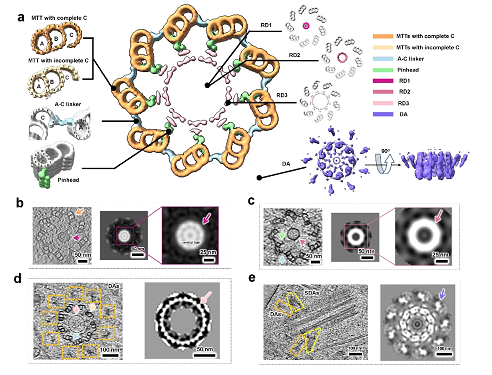
Figure 2. In situ structure of the human centriole. Cross-sectional view of human centriole slices in different local regions, which are reconstructed by STA. MTTs with a complete C-tubule and an incomplete C-tubule are coloured sandy brown and Navajo white, respectively. The A-C linker, pinhead, RD1, RD2, RD3 and DA are coloured light blue, light green, medium violet red, pale violet red, pink and medium slate blue respectively.
The results show that ELI-TriScope will have wide application in future in situ structural biology and in studying the high-resolution ultrastructure of specific events in the cell.
This work entitled "ELI trifocal microscope: a precise system to prepare target cryo-lamellae for in situ cryo-ET study" was published in Nature Methods.
This study was funded by the National Natural Science Foundation of China, Chinese Academy of Sciences, Technological Innovation Program of Chinese Academy of Sciences, Ministry of Science and Technology of the People Republic of China, and Technological Innovation Program of Chinese Academy of Sciences, etc.
Contact
Yun Zhu: University of Chinese Academy of Sciences, E-mail: zhuyun@ibp.ac.cn
Gang Ji: Chinese Academy of Sciences, E-mail: jigang@ibp.ac.cn
Fei Sun: Chinese Academy of Sciences, E-mail: feisun@ibp.ac.cn
Reference
https://www.nature.com/articles/s41592-022-01748-0
Related Articles
CLIEM:https://www.nature.com/articles/s41592-022-01749-z
附件下载: