1997 - 2001 北京大学,学士
2001 - 2007 美国耶鲁大学,博士
2007 - 2013 美国约翰霍普金斯大学,博士后
2013 - 2015 美国约翰霍普金斯大学,研究助理
2016 - 至今 中国科学院生物物理研究所,研究员
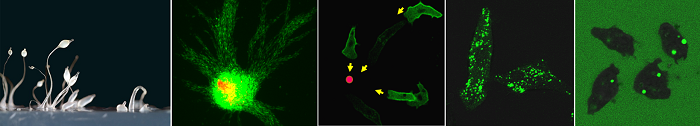
1.细胞巨胞饮的分子机制和生理功能
细胞胞饮,特别是巨胞饮 (macropinocytosis),是细胞摄取营养物质及其它液相大分子的特殊内吞途径,参与免疫反应、病原菌侵袭等重要生物学过程。近年来发现癌细胞可以通过巨胞饮的方式,摄取胞外蛋白质及脂类代谢产物作为营养来源,而阻断胞饮作用可抑制肿瘤生长。作为一种由细胞骨架重排和质膜形变驱动、且伴随大量膜的内化和液体摄入的内吞过程,巨胞饮具有独特的发生机制。此外,巨胞饮过程受到细胞营养状态、环境信号等多种因素的调控,与吞噬和运动等细胞行为之间也存在复杂的调控关系。我们将利用细胞生物学及遗传筛选,系统性鉴定巨胞饮过程的调控基因,阐明它们在胞饮结构起始、形成及成熟过程中的发挥作用的分子机制,并解析这些关键调控基因的病生理功能。
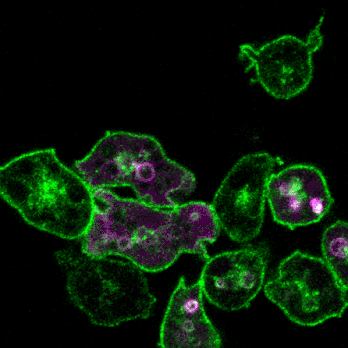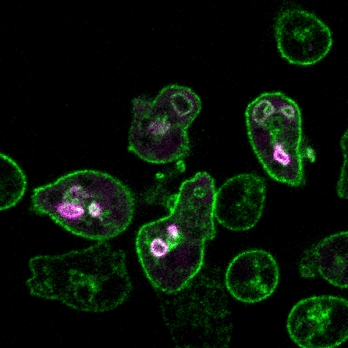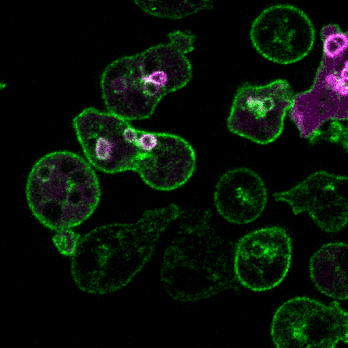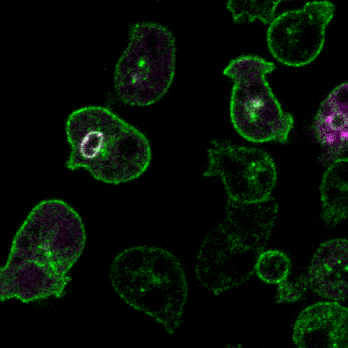

2.细胞定向运动的分子机制和生理功能
细胞运动是进化上保守的生命活动,是胚胎发育、神经元布线、损伤修复及免疫应答等多种生理活动的基础。细胞迁移的异常又与许多人类重大疾病,特别是癌症的发生发展紧密相关。趋化(chemotaxis)是细胞感应外界信号分子时间和空间梯度的定向运动。利用遗传学、细胞生物学、生物化学等多种手段,结合经典模式生物盘基网柄菌(Dictyostelium)和动物细胞模型,我们将致力于研究细胞迁移模式的形成基础、方向感应的工作原理和极性建成的调控机制,以期进一步阐明包括肿瘤转移等复杂生物学过程的分子机理。

1. Hao Y*, Yang Y*, Tu H*, Guo Z*, Chen P, Chao X, Yuan Y, Wang Z, Miao X, Zou S, Li D, Yang Y, Wu C, Li B, Li L#, Cai H#. (2024) A transcription factor complex in Dictyostelium enables adaptive changes in macropinocytosis during the growth-to-development transition. Developmental Cell. 59(5):645-660. (#corresponding)
2. Sun S*, Zhao G*, Jia M*, Jiang Q*, Li S*, Wang H*, Li W*, Wang Y*, Bian X#, Zhao YG#, Huang X#, Yang G#, Cai H#, Pastor-Pareja JC#, Ge L#, Zhang C#, Hu J#. (2024) Stay in touch with the endoplasmic reticulum. Sci China Life Sci. 67(2):230-257. (#corresponding)
3. Li D*, Yang Y*, Lv C, Wang Y, Chao X, Huang J, Singh SP, Yuan Y, Zhang C, Lou J, Gao P, Huang S, Li B, Cai H. (2023) GxcM-Fbp17/RacC-WASP signaling regulates polarized cortex assembly in migrating cells via Arp2/3. Journal of Cell Biology. 222(6):e202208151.
4. Hadwiger JA*,#, Cai H*,#, Aranda RG, Fatima S. (2022) An atypical MAPK regulates translocation of a GATA transcription factor in response to chemoattractant stimulation. Journal of Cell Science. 135(16):jcs260148. (#corresponding)
5. Tu H, Wang Z, Yuan Y, Miao X, Li D, Guo H, Yang Y, Cai H. (2022) The PripA-TbcrA complex-centered Rab GAP cascade facilitates macropinosome maturation in Dictyostelium. Nature Communications. 13(1):1787.
6. Zhang Y*, Tu H*, Hao Y, Li D, Yang Y, Yuan Y, Guo Z, Li L, Wang H#, Cai H#. (2022) Oligopeptide transporter Slc15A modulates macropinocytosis in Dictyostelium by maintaining intracellular nutrient status. Journal of Cell Science. 135(7):jcs259450. (#corresponding) Highlighted in JCS.
7. Li D, Sun F, Yang Y, Tu H, Cai H. (2022) Gradients of PI(4,5)P2 and PI(3,5)P2 jointly participate in shaping the back state of Dictyostelium cells. Frontiers in Cell and Developmental Biology, 10:835185.
8. Yang Y*, Li D*, Chao X, Singh SP, Thomason P, Yan Y, Dong M, Li L, Insall RH, Cai H. (2021) Leep1 interacts with PIP3 and the Scar/WAVE complex to regulate cell migration and macropinocytosis. Journal of Cell Biology. 220(7):e202010096. Highlighted in JCB.
9. Zhuang Y, Zuo D, Tao Y, Cai H#, Li L#. (2020) Laccase3-based extracellular domain provides possible positional information for directing Casparian strip formation in Arabidopsis. PNAS. 117(27):15400-t. (#corresponding)
10. Jiao Z, Cai H#, Long Y, Sirka O, Padmanaban V, Ewarld AJ, Devreotes PN#. (2020) Statin-induced GGPP depletion blocks macropinocytosis and starves cells with oncogenic defects. PNAS. 117(8):4158-4168. (#corresponding)
11. Zhan H, Bhattacharya S, Cai H, Iglesias PA, Huang CH, Devreotes PN. (2020) An Excitable Ras/PI3K/ERK Signaling Network Controls Migration and Oncogenic Transformation in Epithelial Cells. Developmental Cell. 54(5):608-623.
12. Edwards M, Cai H, Abubaker-Sharif B, Long Y, Lampert TJ, Devreotes PN. (2018) Insight from the maximal activation of the signal transduction excitable network in Dictyostelium discoideum. PNAS. 115(16):E3722-3730.
13. Miao Y, Bhattacharya S, Edwards M, Cai H, Inoue T, Iglesias PA, Devreotes PN. (2017) Altering the threshold of an excitable signal transduction network changes cell migratory modes. Nature Cell Biology. 19(4):329-340.
14. Senoo H, Cai H, Wang Y, Sesaki H, Iijima M. (2016) The Novel RacE Binding Protein GflB Sharpens Ras Activity at The Leading Edge of Migrating Cells. Molecular Biology of the Cell. 27(10):1596-1605.
15. Hoeller O*, Toettcher J*, Cai H, Sun Y, Huang CH, Freyre M, Zhao M, Devreotes PN, Weiner O. (2016) Gβ regulates coupling between actin oscillators for cell polarity and directional migration. PLOS Biology. 14(2):e1002381.
16. Ding J, Segarra VA, Chen S, Cai H, Lemmon SK, Ferro-Novick S. (2016) Auxilin facilitates membrane traffic in the early secretory pathway. Molecular Biology of the Cell. 27(1):127-136.
加入生物物理所之前:
17. Santhanam B, Cai H, Devreotes PN, Shaulsky G, and Katoh-Kurasawa M. (2015) The GATA transcription factor GtaC regulates early developmental gene expression dynamics in Dictyostelium. Nature Communications. 6:7551.
18.Cai H, Katoh-Kurasawa M, Muramoto T, Santhanam B, Long Y, Li L, Ueda M, Iglesias PA, Shaulsky G, Devreotes PN. (2014) Nucleocytoplasmic shuttling of a GATA transcription factor functions as a development timer. Science. 343(6177): research article 1249531.
19.Cai H and Devreotes PN. (2011) Moving in the right direction: How eukaryotic cells migrate along chemical gradients. Seminars in Cell and Developmental Biology. 22(8):834-841.
20.Cai H, Das S, Kamimura Y, Long Y, Parent CA, Devreotes PN. (2010) Ras-mediated activation of the TORC2-PKB pathway is critical for chemotaxis. Journal of Cell Biology. 190(2):233-245.
21.Cai H, Reinisch K, Ferro-Novick S. (2007) Coats, Tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Developmental Cell. 12(5):671-682.
22.Cai H, Yu S, Menon S, Cai Y, Lazarova D, Fu C, Reinisch K, Hay JC, Ferro-Novick S. (2007) TRAPPI tethers COPII vesicles by binding the coat subunit Sec23p. Nature. 445(7130):941-944.
23.Cai H, Zhang Y, Pypaert M, Walker L and Ferro-Novick S. (2005) Mutants in trs120 disrupt traffic from the early endosome to the late Golgi. Journal of Cell Biology. 171(5):823-833.
(资料来源:蔡华清研究员,2024-03-19)
蔡华清 博士 研究员 博士生导师
研究方向:细胞运动与胞饮的分子机制及其与疾病的关系
电子邮件:huaqingcai@ibp.ac.cn
电 话:010-64888310
通讯地址:北京市朝阳区大屯路15号(100101)
英文版个人网页:http://english.ibp.cas.cn/sourcedb/rck/EN_xsszmA_G/202005/t20200519_341423.html